
METABIOSYS


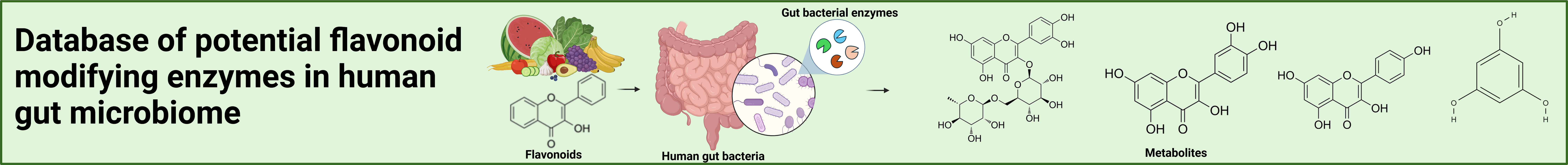
The human gut microbiome plays a crucial role in the metabolism of dietary components by contributing a wide range of enzymatic proteins not encoded by the host genome. Flavonoids, polyphenolic plant secondary metabolites, are commonly found in fruits, vegetables, herbs, seeds, cereals, and beverages. After consumption, some flavonoids are absorbed in the small intestine following O-deglycosylation by epithelial enzymes. In the intestinal epithelium and liver, phase I and phase II transformations of flavonoid aglycons occur. A significant portion of dietary flavonoids (or their phase I/II metabolites) reaches the large intestine, where they are modified by gut bacteria. Various bacterial enzymes are known to carry out flavonoid modifications, including decarboxylation, ring cleavage, reduction, isomerization, and demethylation.
Since a large fraction of the flavonoid-modifying enzymes have been identified and characterized only in recent years, we constructed a comprehensive database of 6,865 potential flavonoid-modifying enzymes (FMEs) from gut bacteria, including 60 biochemically characterized FMEs from published literature and potential FMEs associated with 35 ECs from biochemical databases (BRENDA and KEGG). This could lead to a significant contribution to future biochemical-microbiological investigations on gut bacterial flavonoid transformation.
MetaBioSys lab, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal bypass road, Bhauri, Madhya Pradesh, India (462066).
Copyright © 2022 Indian Institute of Science Education and Research Bhopal, India. All rights reserved.