
METABIOSYS


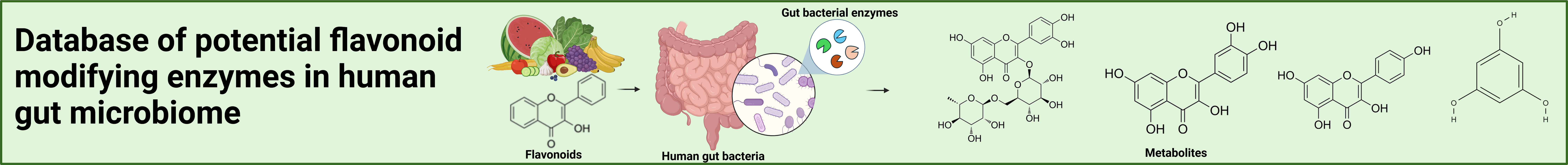
Potential flavonoid-modifying enzymes (FMEs) were retrieved from literature using multiple approaches. A list of hundreds of flavonoid substrates was searched in the BRENDA and KEGG databases. To identify potential FMEs in human gut bacteria, we retrieved 95,303,635 protein sequences (clustered at 100% identity) from the Unified Human Gastrointestinal Protein (UHGP) catalog. The UHGP database, derived from the Unified Human Gastrointestinal Genome catalog v2.0.2 (UHGG), contains 289,232 nonredundant prokaryotic genomes from human gut microbiome. These UHGP sequences were then mapped against bacterial protein sequences from BRENDA and KEGG using Diamond software v2.0.15.153. A total 6,865 bacterial protein sequences were identified to have homologs in the UHGP catalog. These sequences correspond to 35 EC numbers, which are provided below.
MetaBioSys lab, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal bypass road, Bhauri, Madhya Pradesh, India (462066).
Copyright © 2022 Indian Institute of Science Education and Research Bhopal, India. All rights reserved.