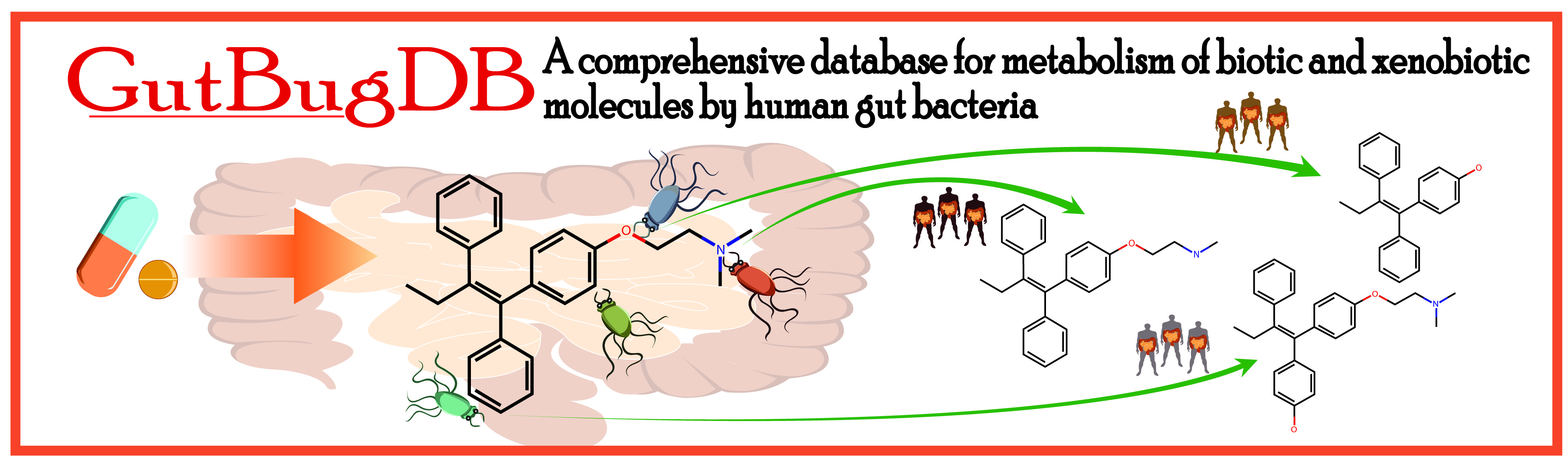
GutBugDB:
GutBugDB database which contains the prediction results for all the FDA-approved drugs. These results were obtained using a hybrid approach developed in our laboratory via implementing machine learning and similarity search approaches. This database was constructed using metabolic proteins from ∼700 gut microbial genomes, 363,872 protein enzymes assigned with their EC numbers (with representative Expasy ID and domains present), and 1399 FDA-approved drugs and nutraceuticals. Gut microbial species are capable of metabolizing a drug molecule with their specific metabolizing enzymes present in the database that can be browsed by the user.
Significance:
Gut microbiota harbors approximately 1000 different bacterial species, hence possessing a vast metabolic potential other than coded by the host genome. Recent reports have shown that some of the drug molecules can be metabolized in the gut lumen by specific gut microbial species. These reports have shifted the focus towards the identification of the mechanism by which gut microbe can metabolize a drug molecule. Most of the orally administered drugs first encounter the gut microbes and their metabolic enzymes prior to their absorption into blood therefore these gut bacterial species play an important role in deciding upon the actual physiological activity of the drug molecule via altering their pharmacokinetic and pharmacodynamic properties. This gut microbial-mediated metabolism can ultimately affect the overall efficacy and toxicity of these drugs. Considering the variation in the gut microbial community between different individuals, thus, this gut microbiota-mediated metabolism of drug molecules can ultimately lead to individual and/or population-specific differences in the drug response.
We have created a database called “GutbugDB” which contains information on gut bacterial species and corresponding metabolic enzymes capable of metabolizing FDA-approved drug molecules. The structural properties of known substrate molecules belonging to particular enzyme class and subclass have been used to train machine learning algorithm for the prediction of enzyme class and subclass, and the same properties have been also used for the similarity search, against the substrate database of particular enzyme subclass, in order to assign exact EC number to new (drug) molecules. Human gut microbes that contain drug metabolizing enzymes are provided in the database along with the representative 3-dimensional structure of the enzyme and the functional domains required for the metabolism. The contribution of specific gut bacterial phylum, genus, and species to the metabolism of FDA-approved drugs has been predicted and included in the database. GutbugDB is repeatedly updated and well-compatible with the integration of new data regarding gut bacterial species and drug molecules. We hope that this database will be useful for the researchers working in the field of gut microbial metabolism and will provide valuable information on the metabolism of all the available drugs.
The flowchart of GutBugDB approach is shown below.

 MetaBioSys
MetaBioSys