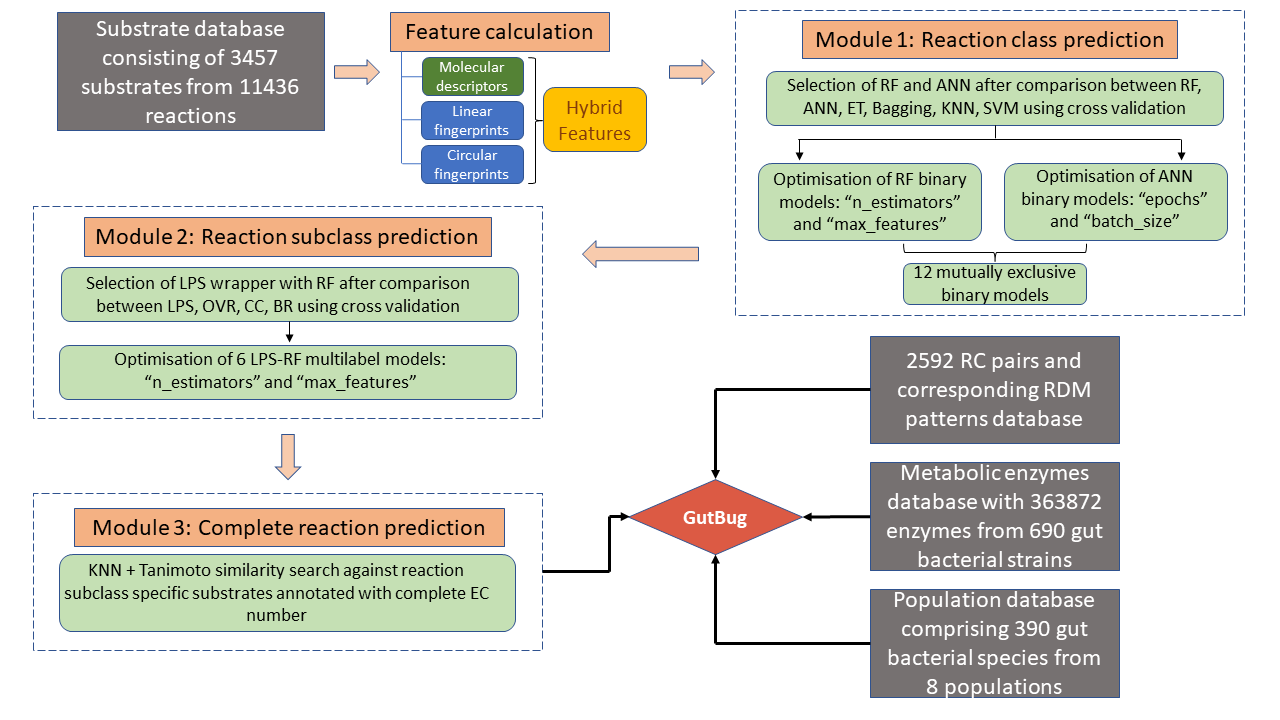
- To identify whether any MAG contains the enzymes predicted by GutBug that are capable of biotransformation of particular substrate, we have provided the pipleine for predicting genes from MAGs using eggNOG-mapper and retrieving GutBug predicted EC numbers for the predicted genes from MAGs.
- The pipeline has been optimised using eggNOG-mapper that uses Diamond for similarity search, Prodigal for gene prediction and utilizes their own annotation database. Using a python script provided, EC numbers predicted by GutBug can be retrieved from MAGs annotated with EC numbers after gene prediction.
- The workflow for utilising GutBug predictions for MAGs analysis is as follows:
- Provide Pubchem ID of a molecule of interest as input into GutBug
- From the Download section at the end of the Result page, download the prediction files for RF and ANN modules as well as download the pipeline for MAGs processing
- Uncompress the MAGs processing pipeline to obtain all the files required for installing eggNOG-mapper and python script for mapping EC numbers predicted by GutBug in the MAGs annotation file that will be generated by eggNOG-mapper
- Instructions regarding installation and usage of eggNOG-mapper are provided in the file named "eggnog_install_and_use.txt". One can also use online version of eggNOG
- For demonstration purpose, prediction results for Zonisamide(Pubchem ID:5734) and MAG annotations file generated by eggNOG-mapper is provided that are used as input for the python script.
- For more information, refer README.txt file.
|
|