



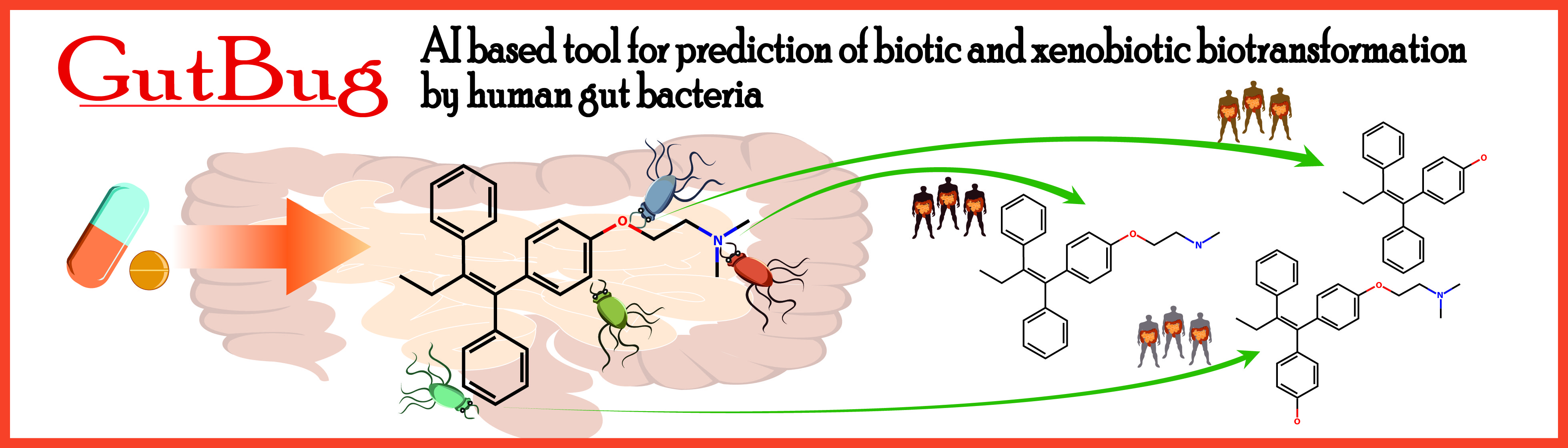
GutBug can predict the human gut bacteria mediated metabolism and biotransformation of biotic and xenobiotic molecules, predicting all possible reactions with reaction centers, corresponding enzymes, gut bacterial strains and abundances for individual bacterial species across different populations.
Human gut microbiome harbours more than 1000 different bacterial species constituting almost 3.3 million unique genes. This provides the gut microbiome immense metabolic potential that can metabolise orally administered xenobiotics/drugs as well as biotic molecules present in our diet. This metabolic potential of gut bacteria can alter pharmacokinetics and pharmacological properties of xenobiotics and deplete key nutrients in diet, leading to individual or population specific variations in drug/diet responses. Large diversity and complexity of the gut microbiome makes any experimental investigations of species-specific metabolism of orally ingested molecules a challenging task. To address this problem, employing machine learning, neural networks and chemoinformatic methods, we developed GutBug. GutBug exploits the promiscuity of metabolic enzymes (an enzyme can potentially bind or metabolize multiple substrates which are structurally similar). GutBug can predict multiple enzymes from multiple gut bacterial strains. GutBug utilises 3,457 enzyme substrates for training and a curated database of 363,872 enzymes from ~700 gut bacterial strains. Molecular properties of all substrates were translated into machine readable features (Descriptors, Fingerprints) and were used for preparing machine learning. GutBug has three modules, for prediction of reaction class, reaction subclass and finally the complete metabolic reaction, following which reaction centres on the query molecule and gut bacterial strains capable of performing that metabolic reactions are identified. Module-1 utilises 6 RF-based and 6 ANN-based binary classifiers (one each for all reaction classes), Module-2 utilises 6 RF-based multilabel models (one each for all reaction classes) and Module-3 utilises KNN plus Tanimoto similarity for prediction of complete metabolic reactions. We have achieved an accuracy of >80% which is very challenging to obtain for a multi-label multi-class classification problem. Thus, GutBug can help predict gut bacteria mediated biotransformation of orally administered molecules. Predictions obtained from GutBug will have valuable impact on pharmacology and drug/diet designing.
GutBug predicts EC number with reaction, bacterial enzymes, gut bacterial strains, and abundance profile of predicted gut bacterial species across different populations for the metabolism of input molecule using the following steps:
MetaBioSys lab, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal bypass road, Bhauri, Madhya Pradesh, India (462066).
Copyright © 2022 Indian Institute of Science Education and Research Bhopal, India. All rights reserved.