ProInflam
Prediction of Proinflamatory epitopes
| Peptide Prediction | Protein Scan | Epitope Mapping | Similarity Search |
|---|
Peptide Prediction |
This page is designed to facilitate user to predict proinflamatory property of peptides. User is allowed to query by pasting one or more peptide sequences in Fasta format or upload a Fasta file as shown below. The Threshold value can be altered from -1.0 to 1.0 with 0.1 step size, to set the stringency of predictions. Click Submit button to view the result page. |
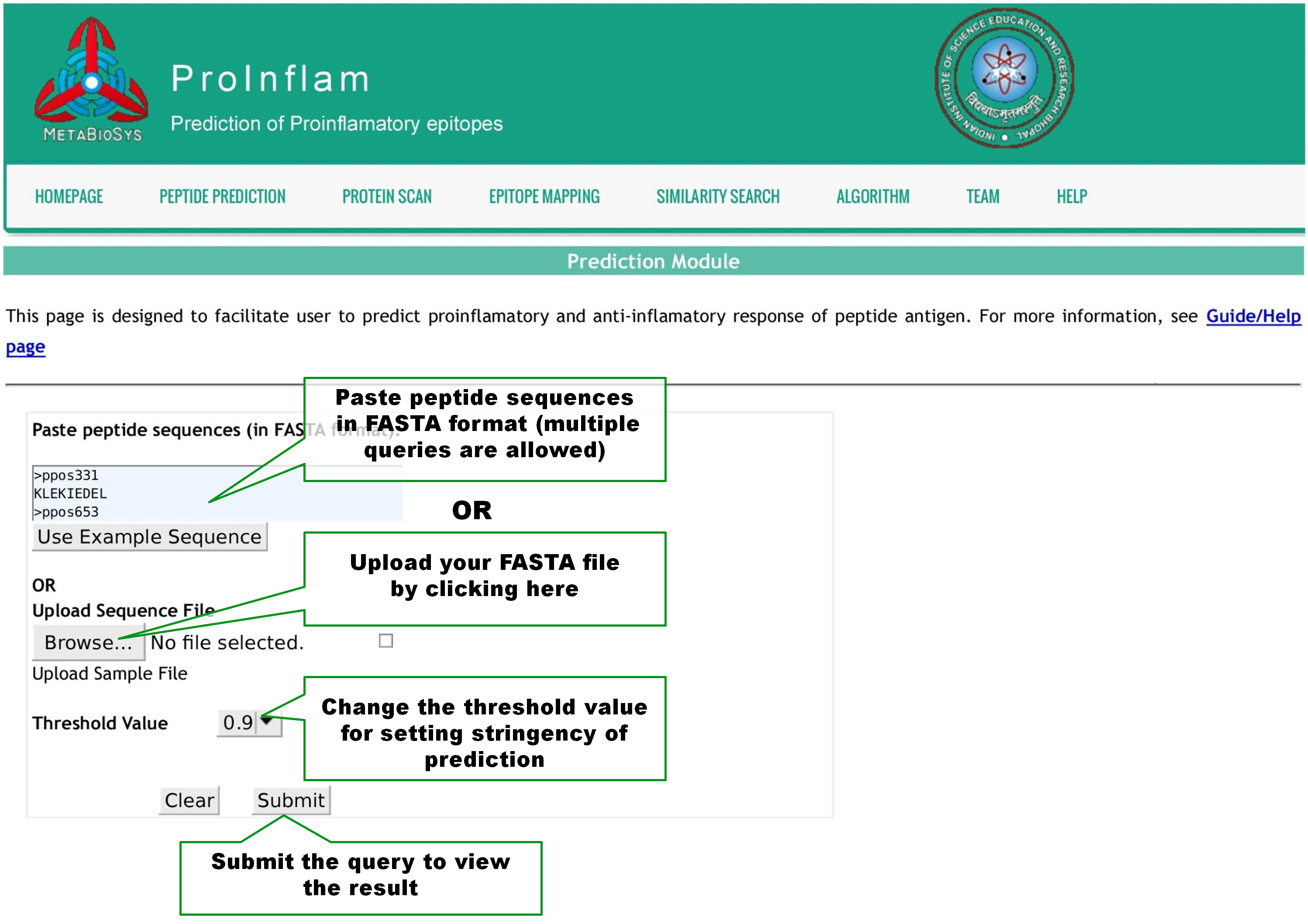
|
In the result page of Peptide prediction, query peptides are listed with scores of prediction and the predicton result(proinflammatory or negative). Results can be downloaded. User can click on a peptide for its virtual screening, a process which predict the proinflammatory property of all of its variants. |
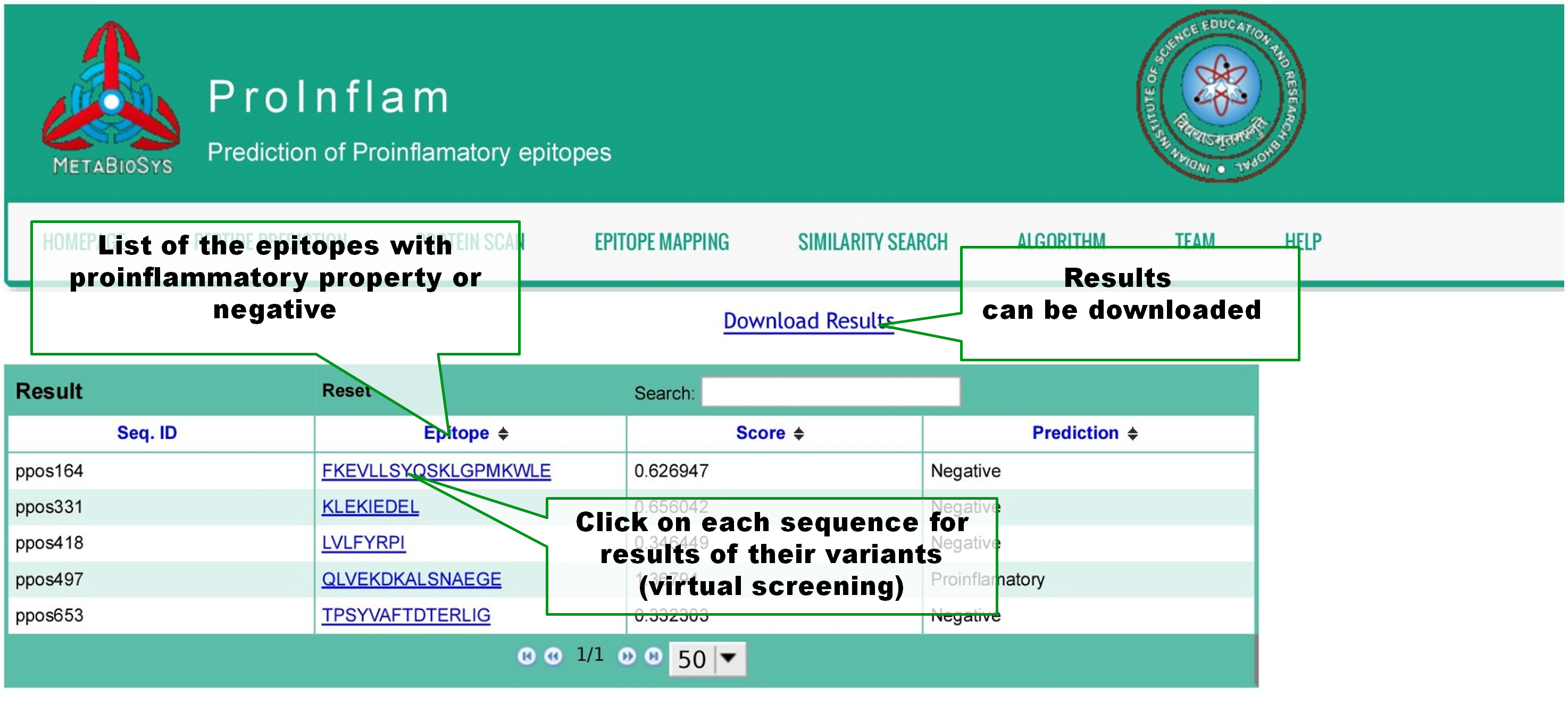 |
Virtual screening will create all the variants of input peptide by substituting each position of it with the all 19 other naturally occuring aminoacids. If 'n' number of aminoacids are present in the input peptide, prediction results of 'n x 19' variant peptides will be shown in tabular format. |
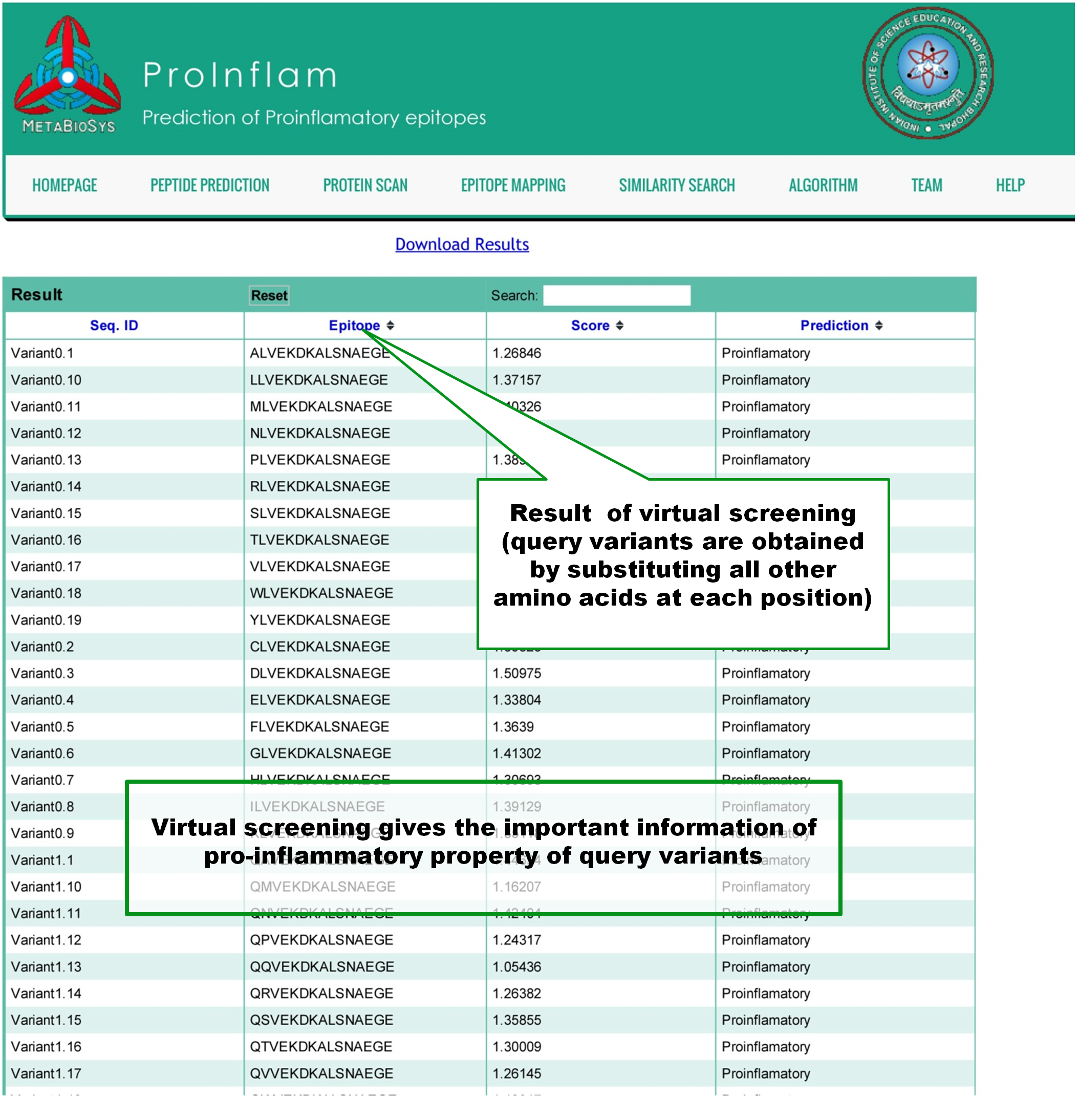 |
Protein Scan |
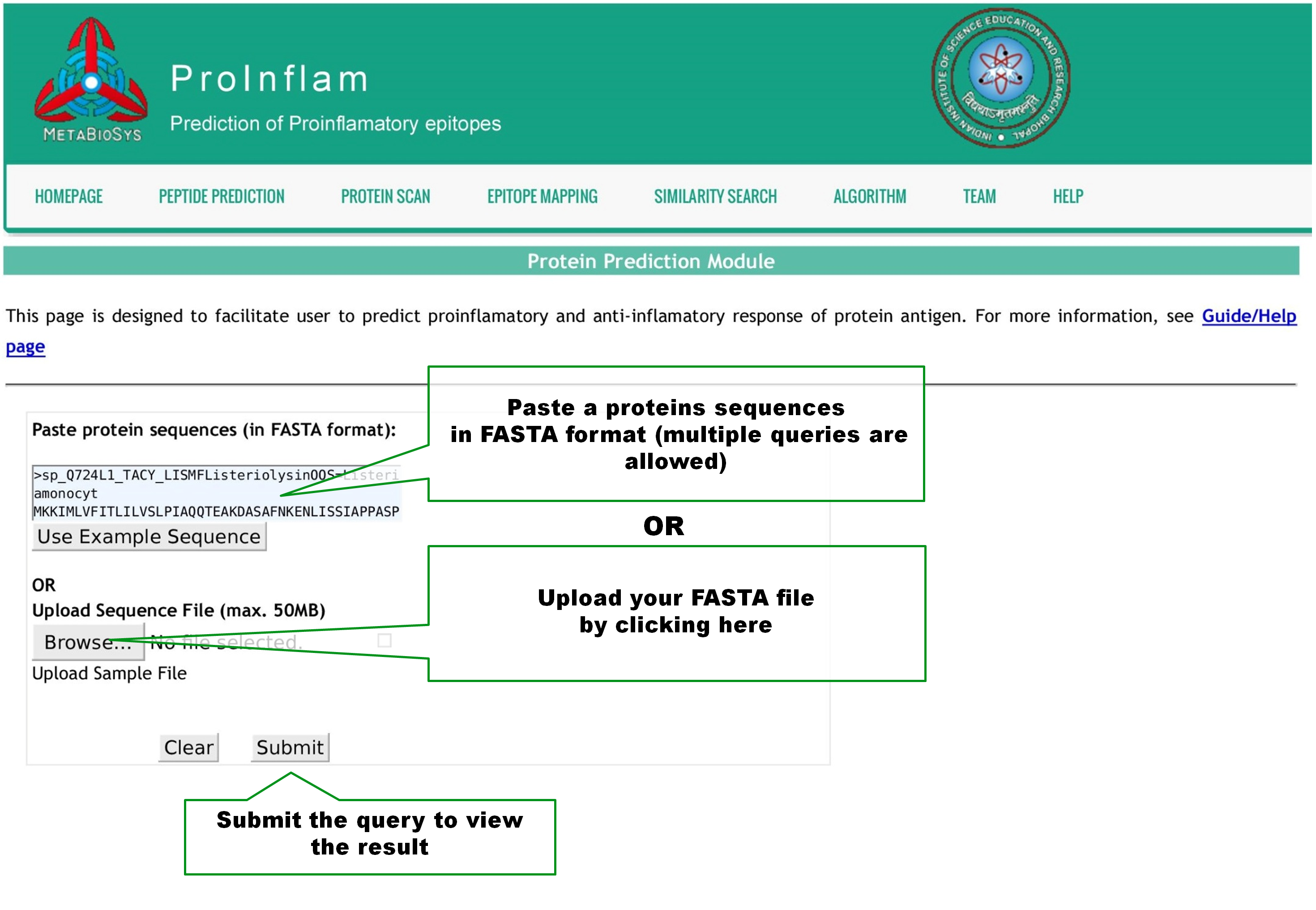
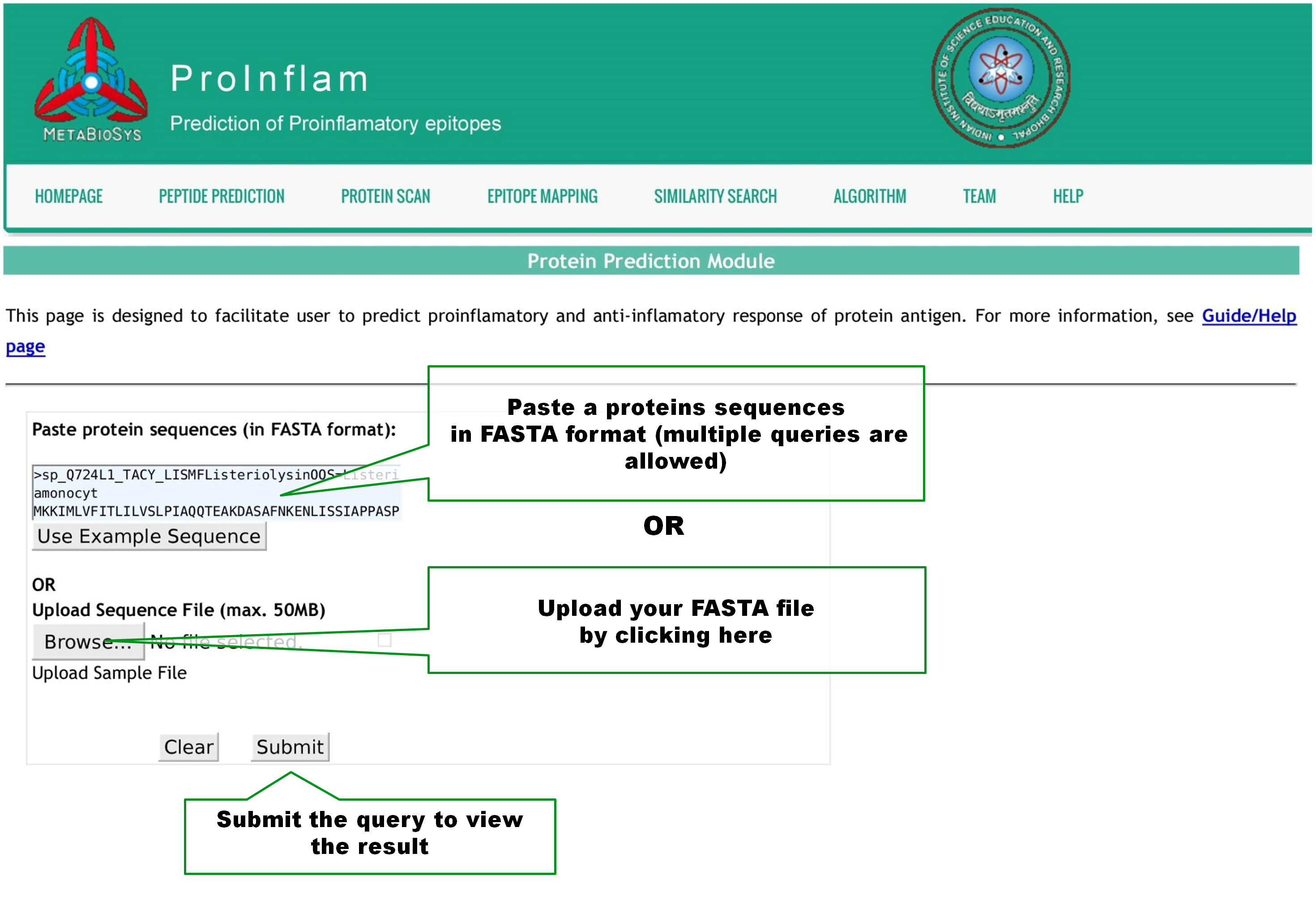
This page is designed to facilitate user to scan the presence of proinflamatory sequences in a protein. Query by pasting one or more protein sequences in Fasta format or upload a Fasta file as shown below. User can change the window length of the peptide sequences, between 4 and 30, used for prediction. The Threshold value can be altered from -1.0 to 1.0 with 0.1 step size, to set the stringency of predictions. Click Submit button to view the result page. |
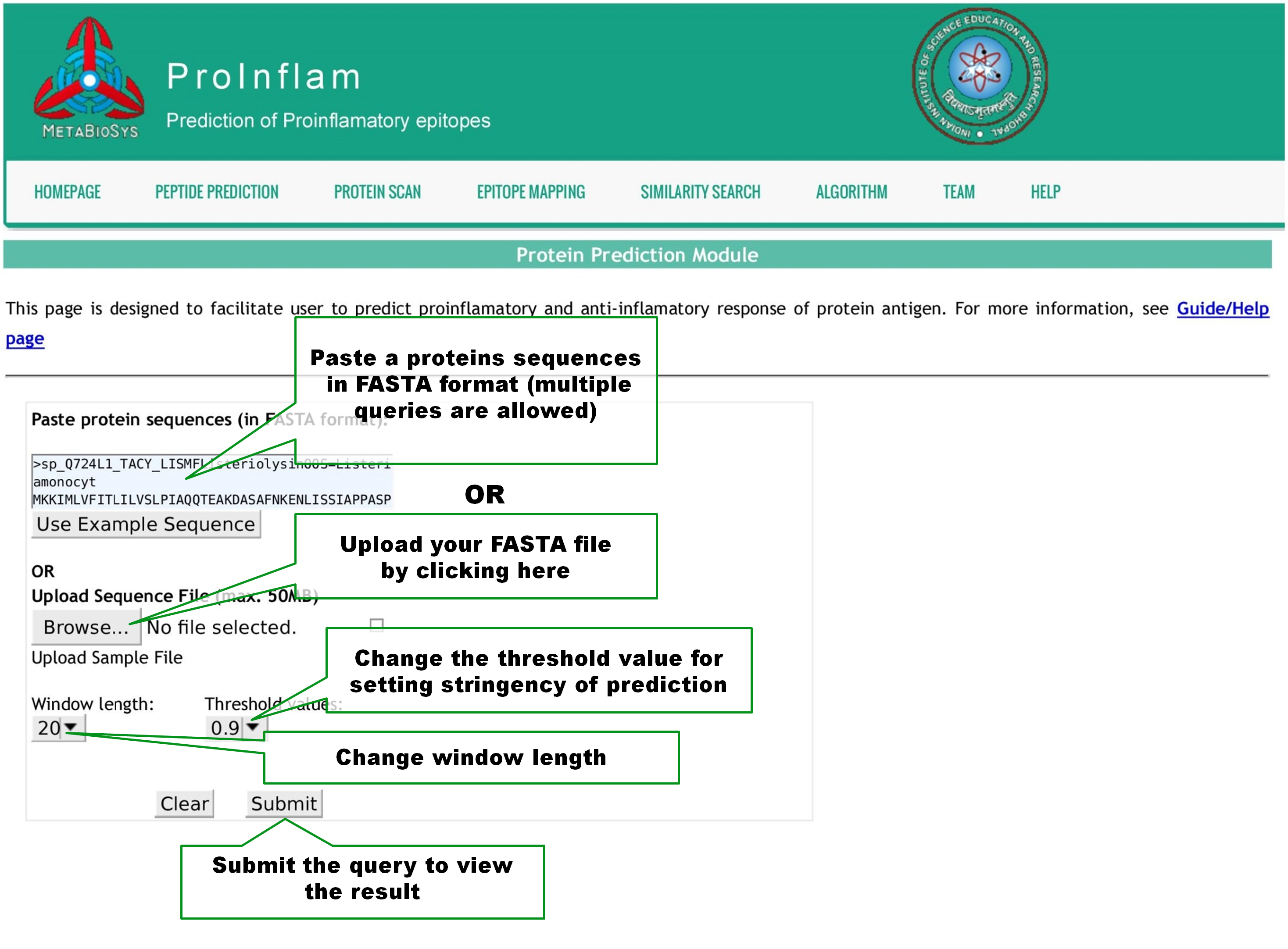 |
In the result page of Protein scan, query peptides are listed with scores of prediction and the predicton result(proinflammatory or negative). Results can be downloaded. User can click on a peptide for its virtual screening, a process which predict the proinflammatory property of all of its variants. |
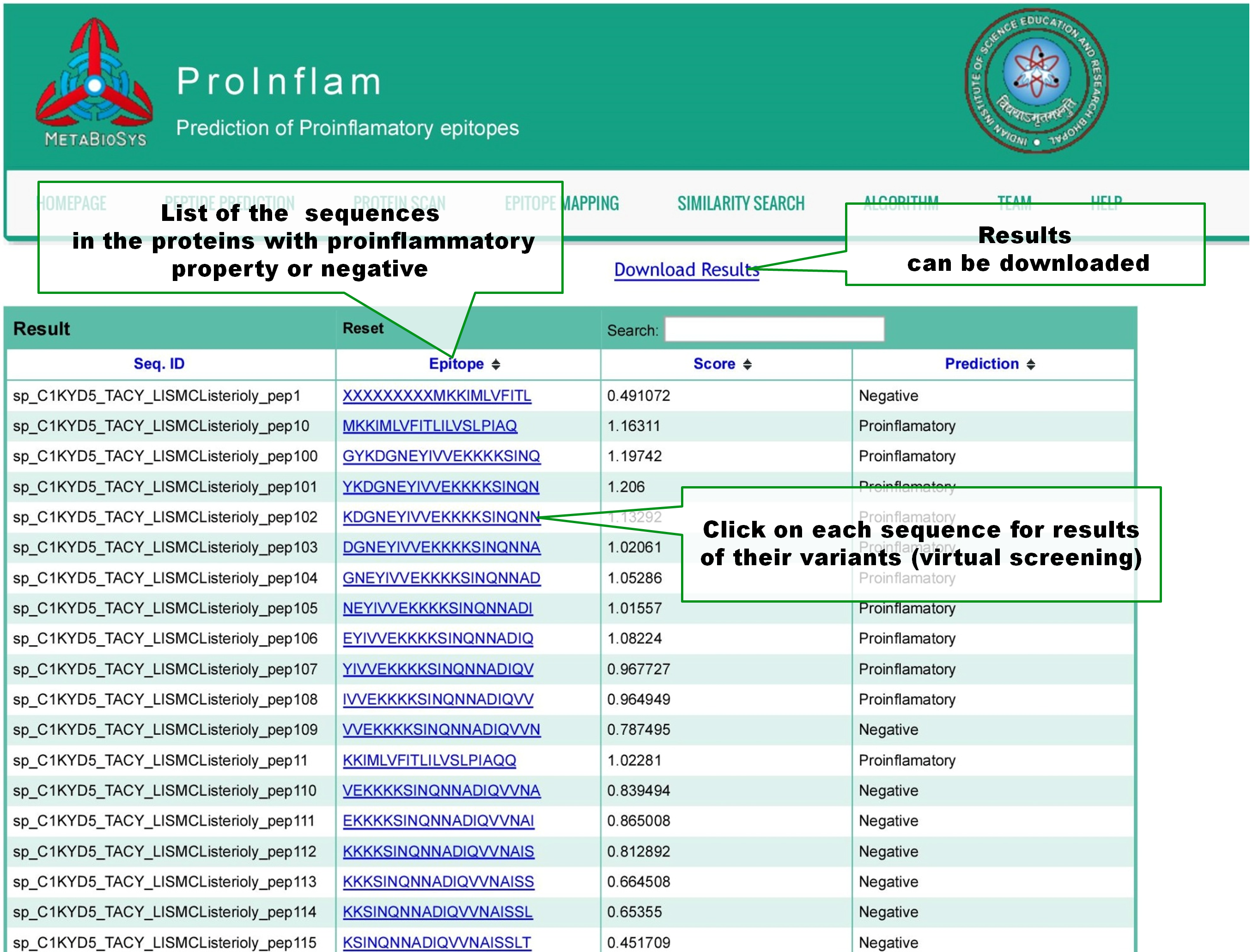 |
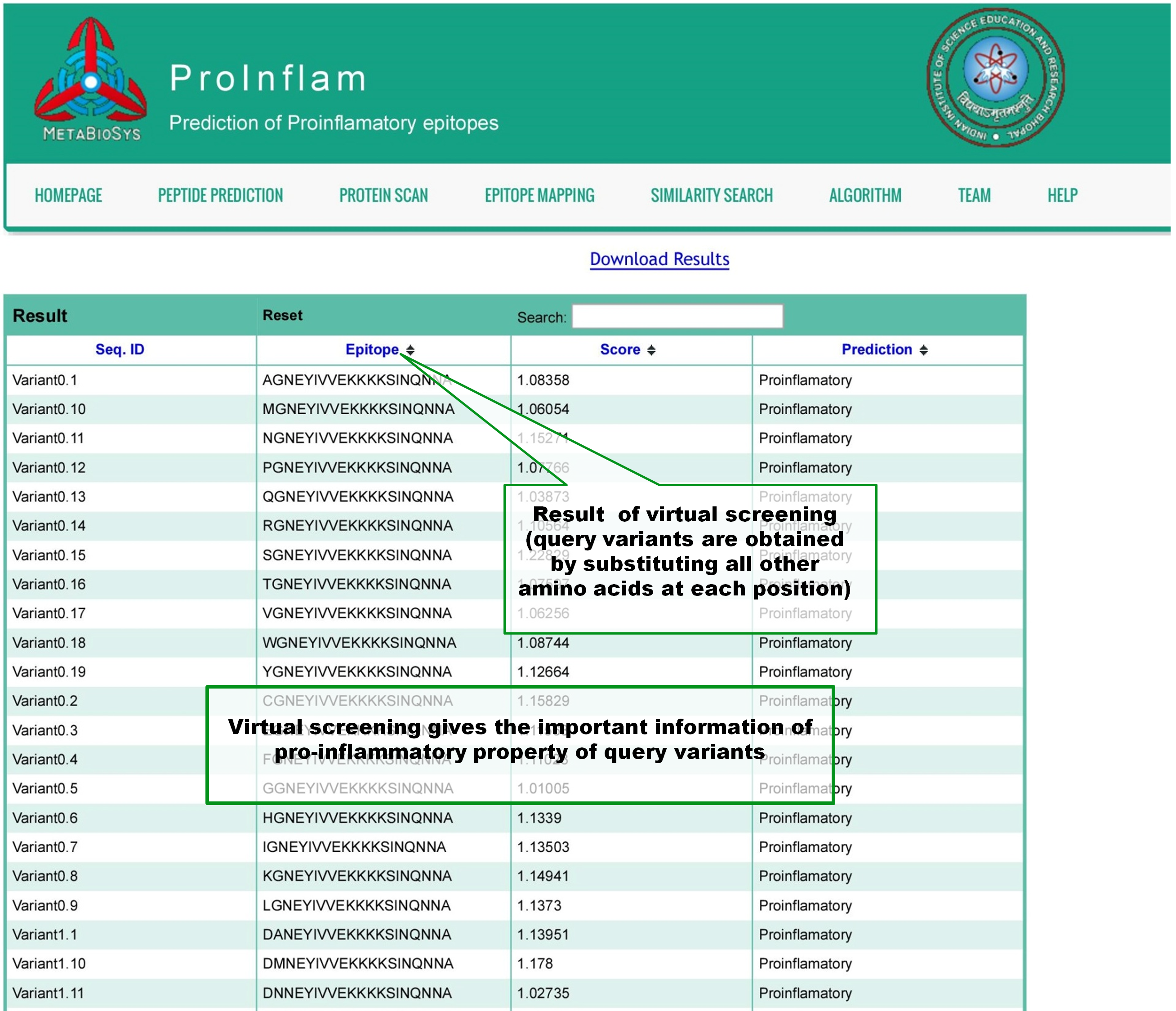
Virtual screening will create all the variants of input peptide by substituting each position of it with the all 19 other naturally occuring aminoacids. If 'n' number of aminoacids are present in the input peptide, prediction results of 'n x 19' variant peptides will be shown in tabular format. |
 |
Epitope Mapping |
This page is designed to facilitate user to map experimentally validated proinflammatory epitopes, into protein/peptide sequences of interest. Query by pasting one or more sequences in Fasta format or upload a Fasta file as shown below. Click Submit button to view the result page. |
 |
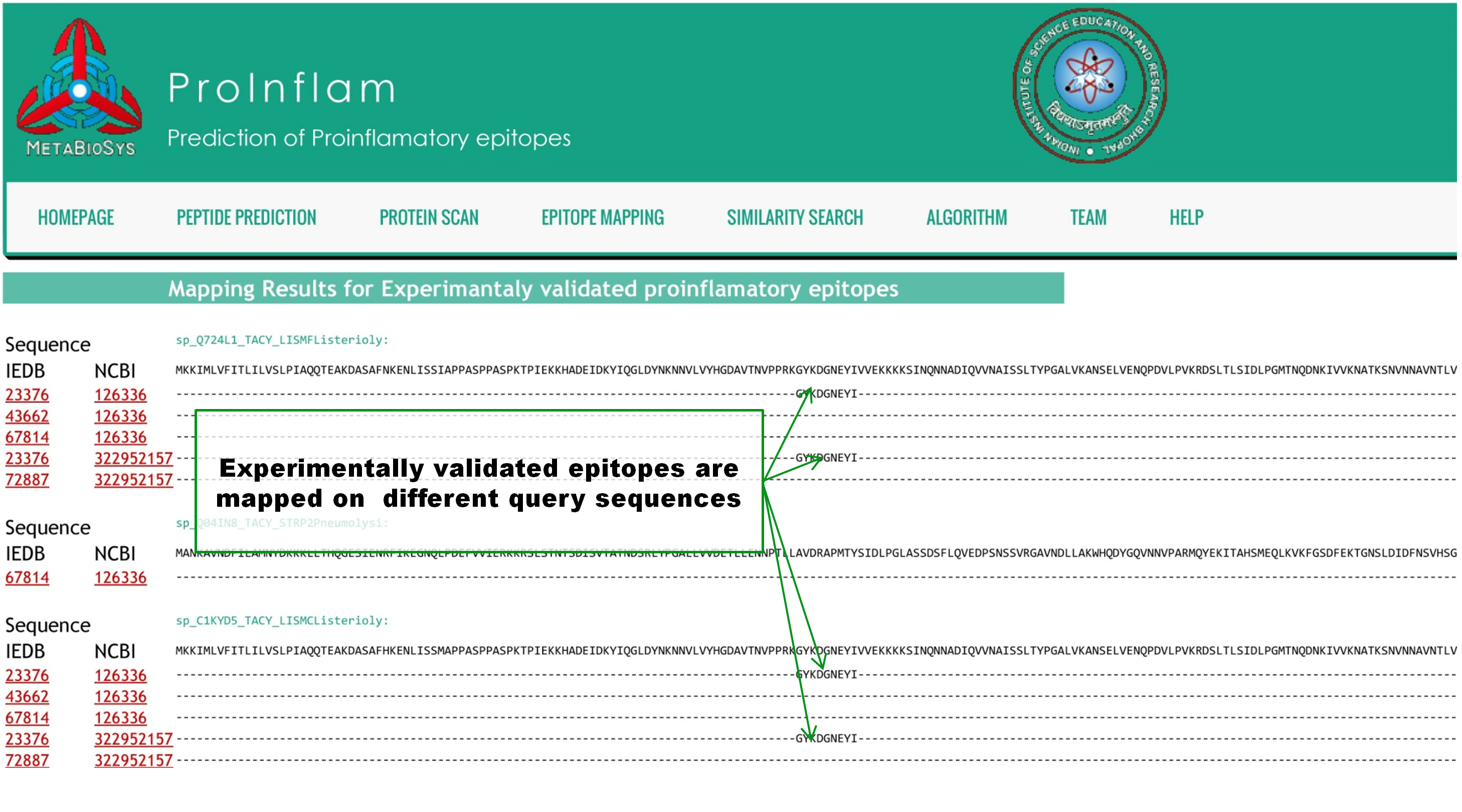
In the result page of Epitope mapping, query sequences are aligned with experimentally validated epitopes from IEDB(Immune Epitope Databse and analysis Sourse) database. IEDB and NCBI IDs of matched epitopes are given with alignments. |
 |
Similarity Search |
This page is designed to facilitate user to do Similarity Search of sequence of interest, against experimantaly validated proinflamatory Epitopes in IEDB(Immune Epitope Databse and analysis Sourse) database. User is allowed to query by pasting one or more peptide sequences in Fasta format or upload a Fasta file as shown below. Click Submit button to view the result page. |
 |
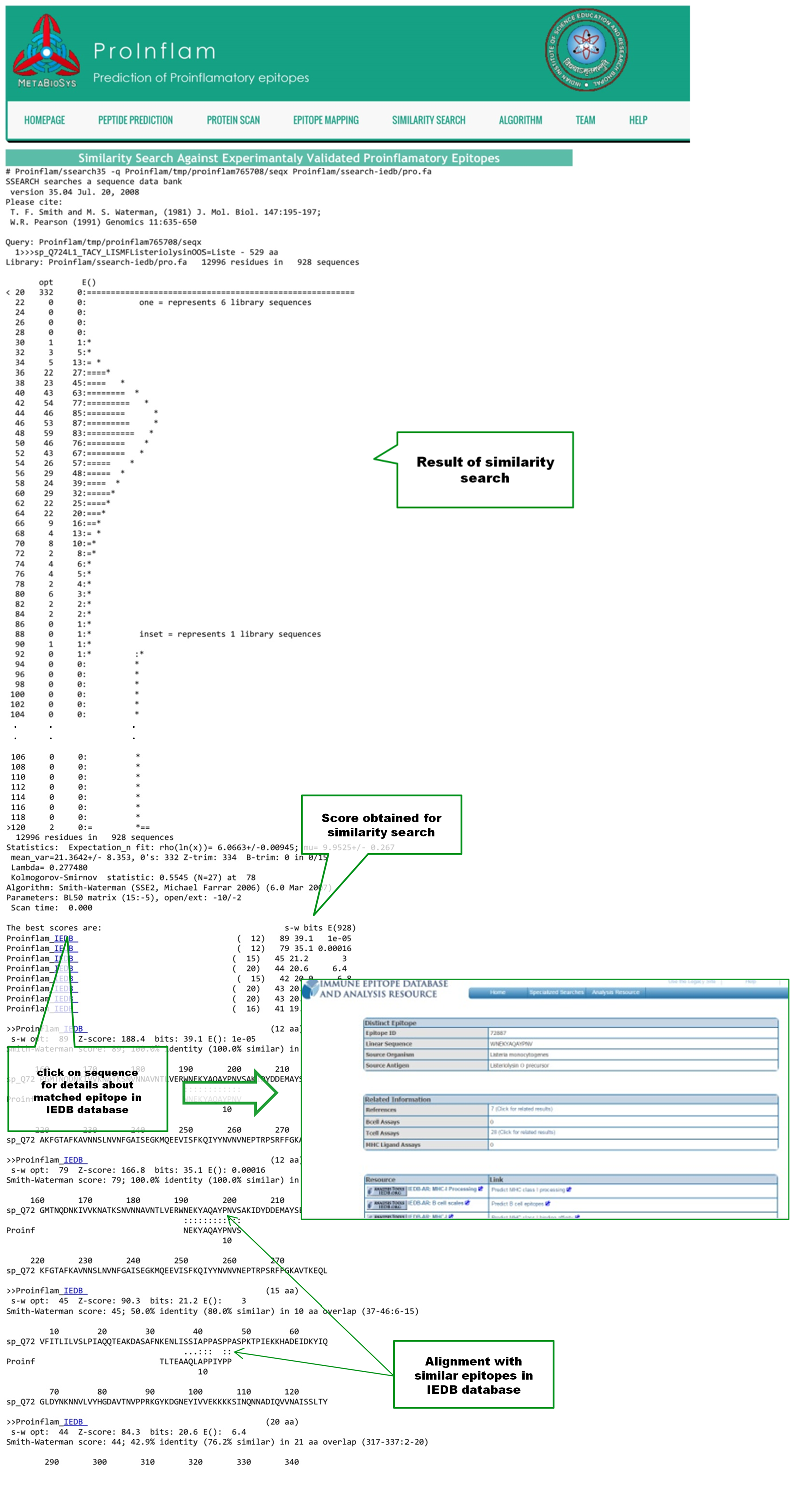
In the result page of Similarity Search, unlike exact epitope mapping, epitopes with similar sequence of query are matched. Scores for all matches are shown. Click on the link to IEDB(Immune Epitope Databse and analysis Sourse) database for viewing the details of matched epitope. Alignment of matched epitopes with the query sequences are also shown |
 |