




This website uses session cookies
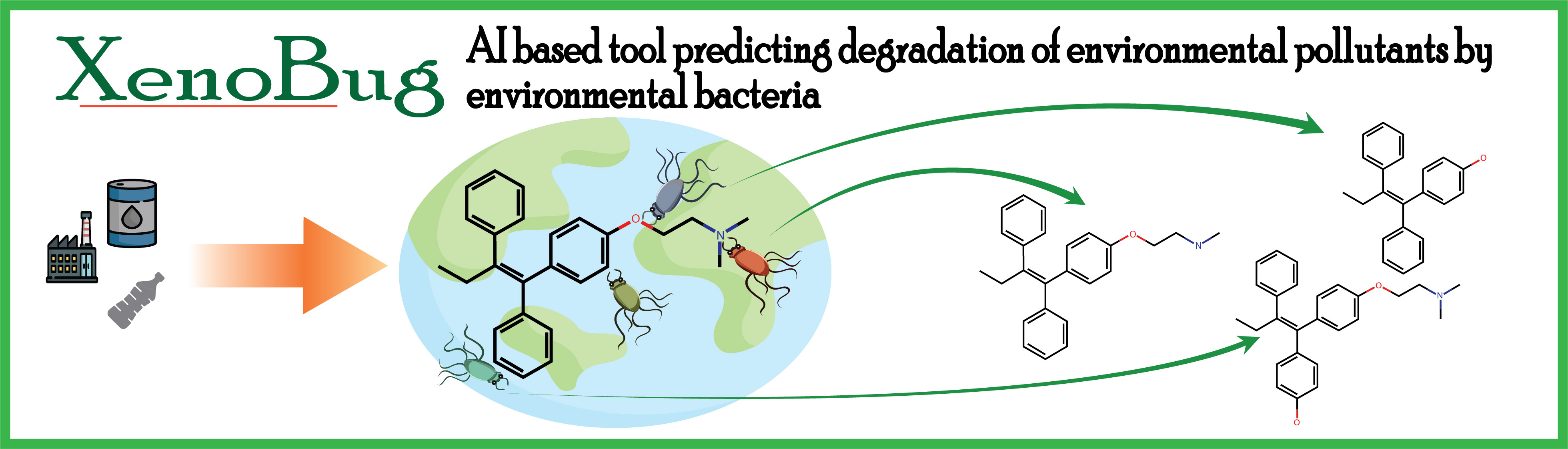
XenoBug can predict the bacterial metabolism and biotransformation of enviornmental pollutants and contaminants, predicting all possible reactions, corresponding enzymes and their sequences from different metagenomic sources. XenoBug is available for free for academic as well as commercial use
Rapid industrialization population growth and modernized agriculture has resulted in excessive usage and accumulation of pesticides, petroleum products and recalcitrant compounds. Accumulation of such toxic and carcinogenic xenobiotics causes environmental pollution causing adverse ecological damage as well as affects human health. Bioremediation strategies that utilise biotransformation or absorption of contaminants using bacterial biochemical activities have better utility for xenobiotic mitigation.
To identify novel bacterial enzymes capable of degrading specific xenobiotics, using machine learning-based predictive approaches to predict enzymes involved in degradation of specific xenobiotics becomes important. Here, we have developed "XenoBug", a machine learning-based tool that predicts EC numbers of bacterial enzymes capable of biodegrading any contaminant of interest and identify the source and sequence of the predicted enzymes in a pre-compiled metagenomic sequence database.
XenoBug predicts EC number with reaction, environmental bacterial enzymes and provides sequence of predicted bacterial enzymes across different metagenomic sources for the metabolism of input molecule using the following steps:
MetaBioSys lab, Indian Institute of Science Education and Research (IISER) Bhopal, Bhopal bypass road, Bhauri, Madhya Pradesh, India (462066).
Copyright © 2023 Indian Institute of Science Education and Research Bhopal, India. All rights reserved.